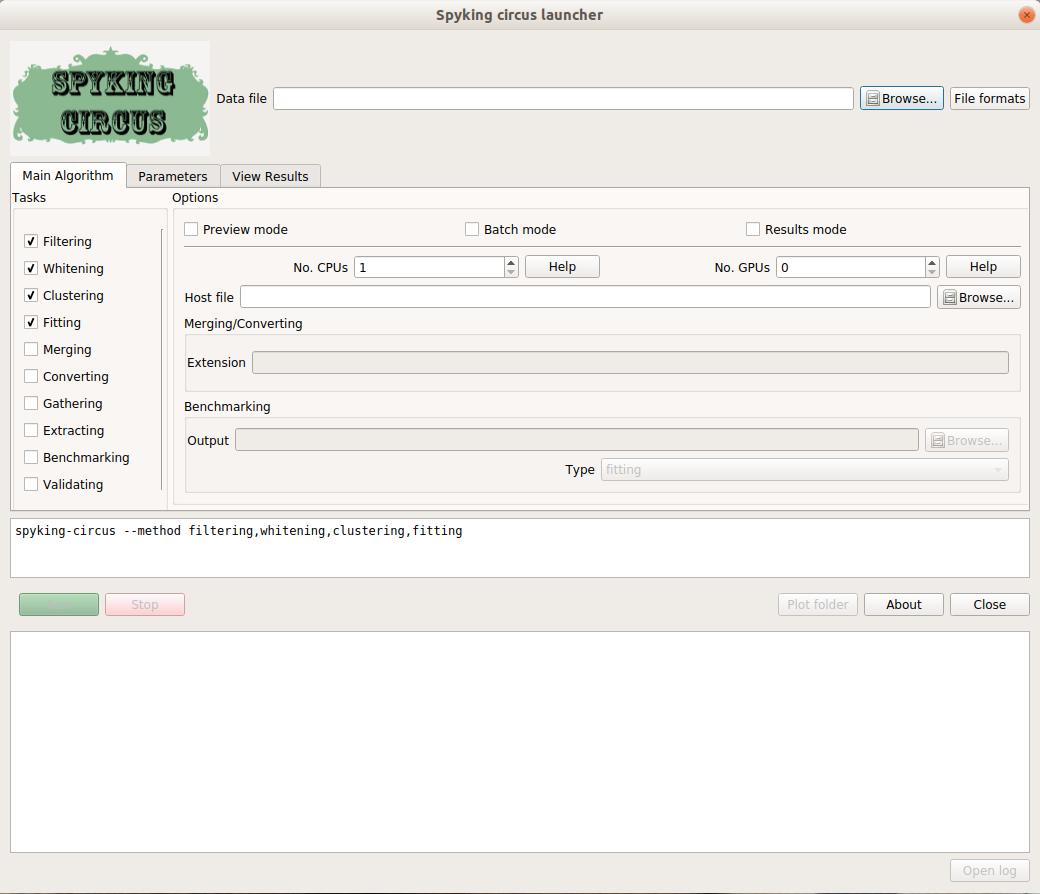
Welcome to the SpyKING CIRCUS’s documentation!¶

The SpyKING CIRCUS is a massively parallel code to perform semi automatic spike sorting on large extra-cellular recordings. Using a smart clustering and a greedy template matching approach, the code can solve the problem of overlapping spikes, and has been tested both for in vitro and in vivo data, from tens of channels to up to 4225 channels. Results are very good, cross-validated on several datasets, and details of the algorithm can be found in the following publication: http://biorxiv.org/content/early/2016/08/04/067843
Warning
We strongly recommend to upgrade to 0.6.xx, as a rare but annoying bug has been found in 0.5.xx while exporting value for post-processing GUIs (MATLAB, phy). The bug is not systematic, depending on the numbers of templates/cores, but it is worth upgrading. A patch will be automatically applied to already sorted results while launching the GUIs, but for phy users, you will need to relaunch the converting step